I'm trying to merge two colorRampPalette schemes to use in leaflet and have been following this nice example. That example works fine but I can't seem to get it to work for my work, reproducible example below. I'm using RdYlGn palette and I want numbers below the threshold to be dark green and numbers above the threshold to more red (skipping some of the inner colors).
For my example my cut-off is nc$PERIMETER < 1.3 so I want numbers under this value to be green and everything above more red (color #FDAE61 onwards).
library(sf)
library(leaflet)
library(RColorBrewer)
#palette im using
palette <- rev(brewer.pal(11, "RdYlGn"))
# [1] "#006837" "#1A9850" "#66BD63" "#A6D96A" "#D9EF8B" "#FFFFBF" "#FEE08B" "#FDAE61" "#F46D43" "#D73027" "#A50026"
previewColors(colorNumeric(palette = palette, domain = 0:10), values = 0:10)
# preparing the shapefile
nc <- st_read(system.file("gpkg/nc.gpkg", package="sf"), quiet = TRUE) %>%
st_transform(st_crs(4326)) %>%
st_cast('POLYGON')
nc
x <- sum(nc$PERIMETER < 1.3)
x # number of values below threshold = 21
### Create an asymmetric color range
## Make vector of colors for values smaller than 1.3 (21 colors)
rc1 <- colorRampPalette(colors = c("#006837", "#1A9850"), space = "Lab")(x) #21
## Make vector of colors for values larger than 1.3
rc2 <- colorRampPalette(colors = c("#FDAE61", "#A50026"), space = "Lab")(length(nc$PERIMETER) - x)
## Combine the two color palettes
rampcols <- c(rc1, rc2)
mypal <- colorNumeric(palette = rampcols, domain = nc$PERIMETER)
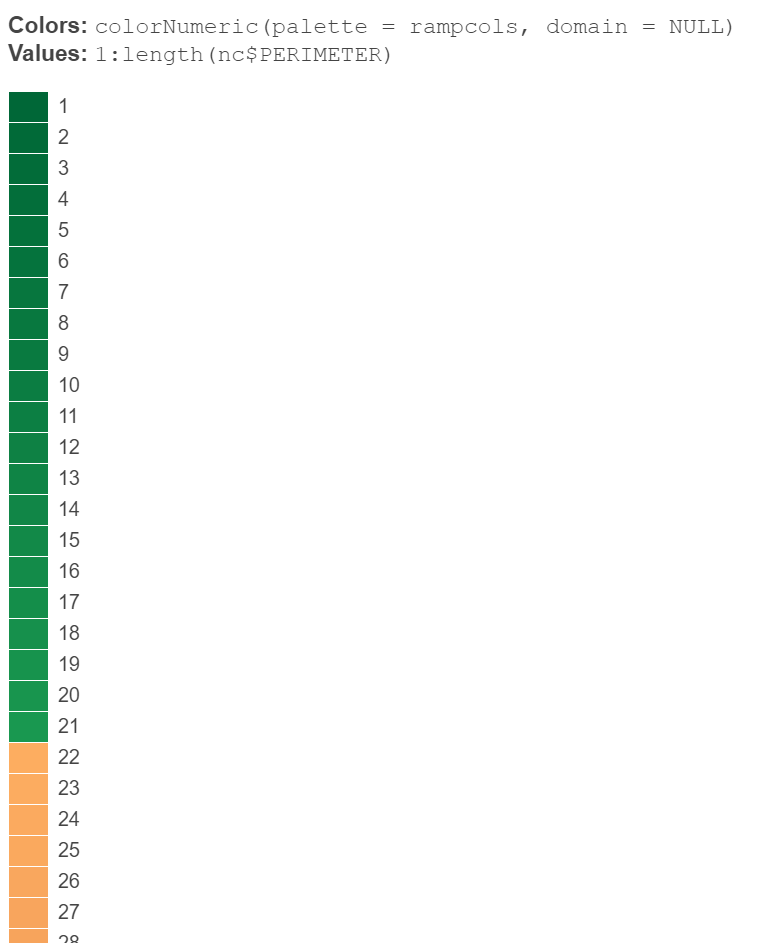
previewColors(colorNumeric(palette = rampcols, domain = NULL), values = 1:length(nc$PERIMETER))
looking at the preview it seems to have worked (21 values under 1.3 should be green):
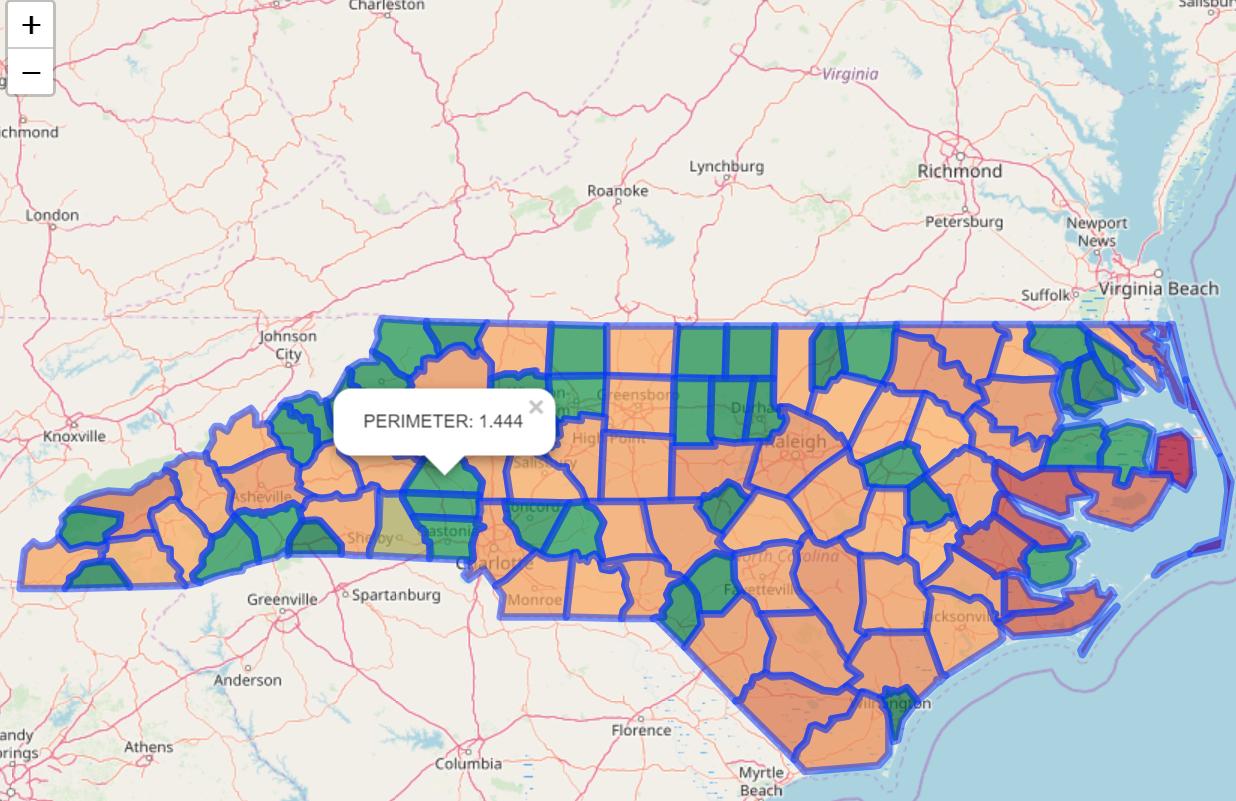
plotting it:
leaflet() %>%
addTiles() %>%
addPolygons(data = nc,
fillOpacity = 0.7,
fillColor = ~mypal(PERIMETER),
popup = paste("PERIMETER: ", nc$PERIMETER) )
plots ok but doesn't give the right color, the one highlighted is above the threshold (1.3) and so shouldn't be green but it is:
I thought the way I was creating the palettes was wrong but the preview seems to suggest I've done it right?
anyone have any ideas? thanks