Delaunay/Voronoi methods can be used for this problem, since the medial axis is a sub-graph of the Voronoi graph
(see for example this paper by Attali, Boissonnat and Edelsbrunner).
In the following I will demonstrate the methods on an example of points sampled from a quarter torus surface with small radius 10 and large radius 100 (the medial path/skeleton starts at point (100, 0, 0) and ends at (0, 100, 0)).
The Voronoi graph is the dual of the 3D Delaunay tetrahedralization (I will from now on use the term triangulation for this).
Computing the Delaunay triangulation can be done using scipy's scipy.spatial.Delaunay package.
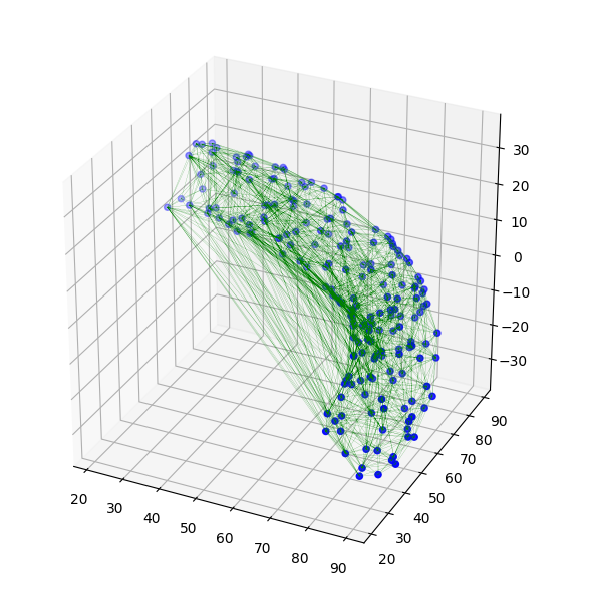
Below is a figure of the sample points (200 in this example) and their full Delaunay triangulation
(the triangulation was plotted with the function from here).
![full triangulation]()
The Voronoi vertex corresponding to a Delaunay tetrahedron is the center of the circumscribing sphere of the tetrahedron.
The following is a function that computes these Delaunay centers, it is an extension to the 2D function from my previous answer here.
def compute_delaunay_tetra_circumcenters(dt):
"""
Compute the centers of the circumscribing circle of each tetrahedron in the Delaunay triangulation.
:param dt: the Delaunay triangulation
:return: array of xyz points
"""
simp_pts = dt.points[dt.simplices]
# (n, 4, 3) array of tetrahedra points where simp_pts[i, j, :] holds the j'th 3D point (of four) of the i'th tetrahedron
assert simp_pts.shape[1] == 4 and simp_pts.shape[2] == 3
# finding the circumcenter (x, y, z) of a simplex defined by four points:
# (x-x0)**2 + (y-y0)**2 + (z-z0)**2 = (x-x1)**2 + (y-y1)**2 + (z-z1)**2
# (x-x0)**2 + (y-y0)**2 + (z-z0)**2 = (x-x2)**2 + (y-y2)**2 + (z-z2)**2
# (x-x0)**2 + (y-y0)**2 + (z-z0)**2 = (x-x3)**2 + (y-y3)**2 + (z-z3)**2
# becomes three linear equations (squares are canceled):
# 2(x1-x0)*x + 2(y1-y0)*y + 2(z1-z0)*y = (x1**2 + y1**2 + z1**2) - (x0**2 + y0**2 + z0**2)
# 2(x2-x0)*x + 2(y2-y0)*y + 2(z2-z0)*y = (x2**2 + y2**2 + z2**2) - (x0**2 + y0**2 + z0**2)
# 2(x3-x0)*x + 2(y3-y0)*y + 2(z3-z0)*y = (x3**2 + y3**2 + z3**2) - (x0**2 + y0**2 + z0**2)
# building the 3x3 matrix of the linear equations
a = 2 * (simp_pts[:, 1, 0] - simp_pts[:, 0, 0])
b = 2 * (simp_pts[:, 1, 1] - simp_pts[:, 0, 1])
c = 2 * (simp_pts[:, 1, 2] - simp_pts[:, 0, 2])
d = 2 * (simp_pts[:, 2, 0] - simp_pts[:, 0, 0])
e = 2 * (simp_pts[:, 2, 1] - simp_pts[:, 0, 1])
f = 2 * (simp_pts[:, 2, 2] - simp_pts[:, 0, 2])
g = 2 * (simp_pts[:, 3, 0] - simp_pts[:, 0, 0])
h = 2 * (simp_pts[:, 3, 1] - simp_pts[:, 0, 1])
i = 2 * (simp_pts[:, 3, 2] - simp_pts[:, 0, 2])
v1 = (simp_pts[:, 1, 0] ** 2 + simp_pts[:, 1, 1] ** 2 + simp_pts[:, 1, 2] ** 2) - (simp_pts[:, 0, 0] ** 2 + simp_pts[:, 0, 1] ** 2 + simp_pts[:, 0, 2] ** 2)
v2 = (simp_pts[:, 2, 0] ** 2 + simp_pts[:, 2, 1] ** 2 + simp_pts[:, 2, 2] ** 2) - (simp_pts[:, 0, 0] ** 2 + simp_pts[:, 0, 1] ** 2 + simp_pts[:, 0, 2] ** 2)
v3 = (simp_pts[:, 3, 0] ** 2 + simp_pts[:, 3, 1] ** 2 + simp_pts[:, 3, 2] ** 2) - (simp_pts[:, 0, 0] ** 2 + simp_pts[:, 0, 1] ** 2 + simp_pts[:, 0, 2] ** 2)
# solve a 3x3 system by inversion (see https://en.wikipedia.org/wiki/Invertible_matrix#Inversion_of_3_%C3%97_3_matrices)
A = e*i - f*h
B = -(d*i - f*g)
C = d*h - e*g
D = -(b*i - c*h)
E = a*i - c*g
F = -(a*h - b*g)
G = b*f - c*e
H = -(a*f - c*d)
I = a*e - b*d
det = a*A + b*B + c*C
# multiplying inv*[v1, v2, v3] to get solution point (x, y, z)
x = (A*v1 + D*v2 + G*v3) / det
y = (B*v1 + E*v2 + H*v3) / det
z = (C*v1 + F*v2 + I*v3) / det
return (np.vstack((x, y, z))).T
We would like to filter out tetrahedra that are outside of the original surface (see for example the long tetrahedra in the figure above).
This might be done by testing the tetrahedra against the original surface.
However, a simpler way, which is very suited for the input of a tube/pipe surface is
to filter out tetrahedra that have a large circumscribing radius.
This is what the alpha-shape algorithm does.
This is easily done within our context since the radius is just the distance between the center and any of the tetrahedron points.
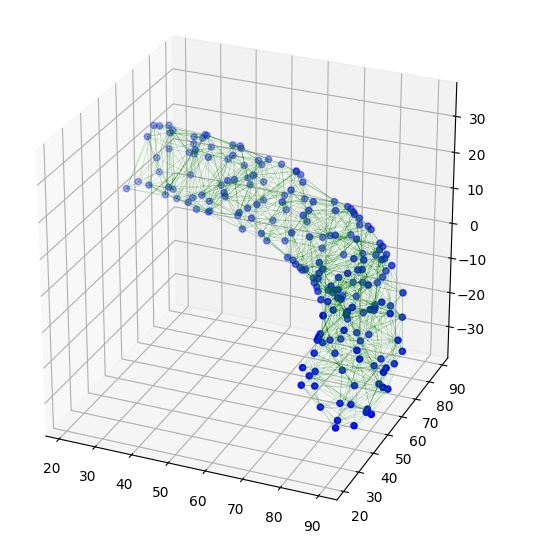
The following figure shows the Delaunay triangulation after filtering out tetrahedra with radius greater than 20.
![3d alpha-shape]()
We can now use these building blocks to build the Voronoi sub-graph of the tetrahedra that pass the radius condition.
The function below does this using the connectivity information in the Delaunay triangulation to construct the Voronoi sub-graph, represented as an edge list.
def compute_voronoi_vertices_and_edges(points, r_thresh=np.inf):
"""
Compute (finite) Voronoi edges and vertices of a set of points.
:param points: input points.
:param r_thresh: radius value for filtering out vertices corresponding to
Delaunay tetrahedrons with large radii of circumscribing sphere (alpha-shape condition).
:return: array of xyz Voronoi vertex points and an edge list.
"""
dt = Delaunay(points)
xyz_centers = compute_delaunay_tetra_circumcenters(dt)
# filtering tetrahedrons that have radius > thresh
simp_pts_0 = dt.points[dt.simplices[:, 0]]
radii = np.linalg.norm(xyz_centers - simp_pts_0, axis=1)
is_in = radii < r_thresh
# build an edge list from (filtered) tetrahedrons neighbor relations
edge_lst = []
for i in range(len(dt.neighbors)):
if not is_in[i]:
continue # i is an outside tetra
for j in dt.neighbors[i]:
if j != -1 and is_in[j]:
edge_lst.append((i, j))
return xyz_centers, edge_lst
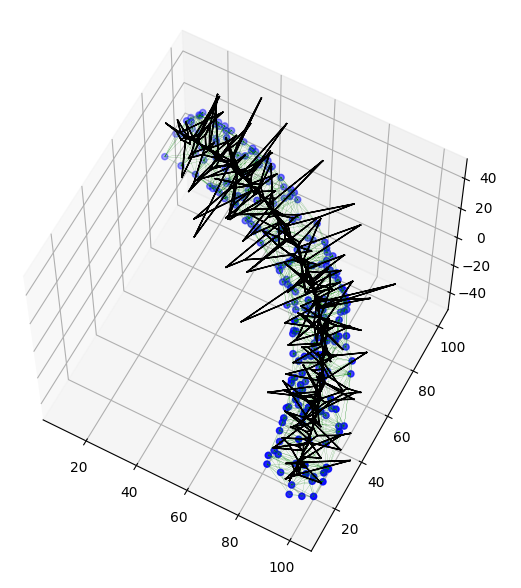
The result is still not sufficient as can be seen in the figure below, where the sub-graph edges are the black line segments.
The reason is that 3D Delaunay triangulations are notorious for having thin tetrahedra
(so-called slivers, needles and caps in this paper by Shewchuk),
which cause the outer "spikes" in the results.
![Voronoi sub-graph]()
While there are general methods to remove these unwanted spikes
(see, for example, Amenta and Bern),
in the context of a tube surface there is a simple solution.
The path we are looking for can be computed as the shortest Euclidean path in the graph starting at the closest point to the start of the tube and ending at the closest point to the end.
The following code does this using a networkx graph with weights set to the lengths of the edges.
# get closest vertex to start and end points
xyz_centers, edge_lst = compute_voronoi_vertices_and_edges(pts, r_thresh=20.)
kdt = cKDTree(xyz_centers)
dist0, idx0 = kdt.query(np.array([100., 0, 0]))
dist1, idx1 = kdt.query(np.array([0, 100., 0]))
# compute shortest weighted path
edge_lengths = [np.linalg.norm(xyz_centers[e[0], :] - xyz_centers[e[1], :]) for e in edge_lst]
g = nx.Graph((i, j, {'weight': dist}) for (i, j), dist in zip(edge_lst, edge_lengths))
path_s = nx.shortest_path(g,source=idx0,target=idx1, weight='weight')
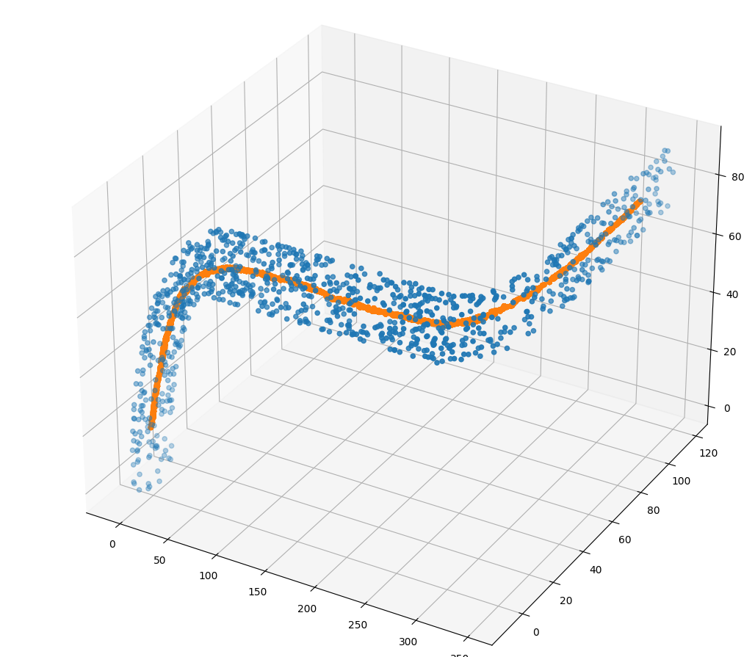
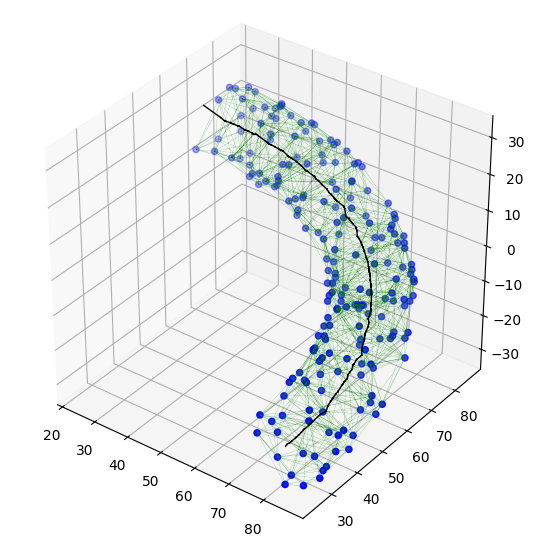
The figure below shows the results for the original 200 points.
![final path for 200 points]()
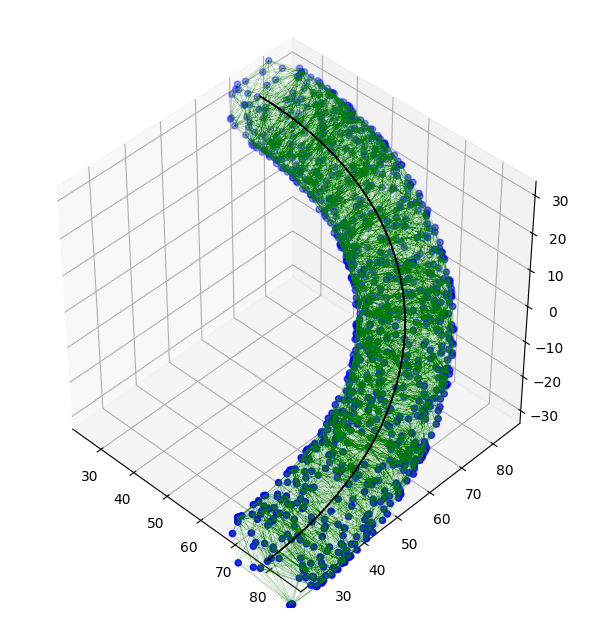
And this is the results for a denser sample of 1000 points.
![final path for 1000 points]()
Now you can pass an approximating spline - interpolating or least-square fit, through the path points.
You can use scipy.interpolate.UnivariateSpline as suggested in the link
or scipy.interpolate.splrep as done here or any other standard spline implementation.